Please select one genome and start searching
Glycine max
978MCultivar Soybean
Williams 82 a2v1
Glycine soja
950MWild Soybean
W05 a1v1
Glycine max
978MCultivar Soybean
Williams 82 a4v1
Glycine max
978MCultivar Soybean
ZH13 v2
Browser Compatibility
For a better experience and fully utilize the features of datahub, we suggest to use the most up-to-date version of the modern browsers. The website was tested on the latest Google Chrome, Firefox, MS Edge and Safari.
Release Note
AoE DataHub updates 07/12/2021
- User now could extract transcript sequences in batches in SeqExtractor
AoE DataHub updates 11/11/2021
- Gene expression could be accessed as barplot in gene search tab
AoE DataHub updates 10/10/2021
- Integrated miRNA expression as heatmap to the gene search result, 8 datasets for Gmax_a4v1 were added.
- Various genomic datasets were added to Jbrowse section, updated list could be viewed here.
AoE DataHub updates 30/07/2021
- Integrated annotation from UniProt/Swiss-Prot database
- Added support for illustrating InterProScan discovered domain result
AoE DataHub updates 16/07/2021
- Documents were online
AoE DataHub updates 29/06/2021
- Synteny Blocks among genomes were analyzed by MCScan and visualization section was integrated.
AoE DataHub updates 22/05/2021
- Flanking sequence could be extracted from gene search tabs.
AoE DataHub 0.0.4 released 18/05/2021
- Added primer design function with primer3 as a backend.
- REBASE (20210430) was integrated.
AoE DataHub 0.0.3 released 15/01/2021
- All the data of four genomes were populated.
- Jbrowse content was updated for new added genomes.
- All genes were reannotated with latest version of interproscan 5.48-83.0
AoE DataHub 0.0.2 released 10/01/2021
- Sequence Extractor function added.
- BLAST function improved, much faster than before.
- Application UI modified.
AoE DataHub 0.0.1 released 24/11/2020
- This is the testing version of prototype, and this section is for release content testing only. For prototype, we added data for W05 only, the BLAST function and Jbrowse was integrated to the shiny dashboard app.
Instructions:
- Choose Reference genome from drop-down menu
-
Select DataSet Type to search against different resource:
- Ref Genome: Genomic sequences
- Ref Transcript: Transcript cDNA sequences
- Ref CDS: Transcript CDS sequences
- Ref Proteome: Transcript protein amino acid sequences
-
Choose proper BLAST program:
- megablast is optimized for comparison of very similiar sequences, e.g. intraspecies paralogues
- blastn is better suited for interspecies comparisons with a shorter word-size
- tblastn for a standard protein-translated nucleotide comparison
- blastn-short is optimized for sequences less than 30 nucleotides, e.g. primers
- Set e-value cutoff, the default one used here is 0.01, which is relaxed to include more hits
Note: Try "Structural Variant Inspector" with our demo data: enter 'Gsoja.W05/Wm82_pacbio_hifi.ref_W05.SV.demo.vcf' into the "url" box and select "Glycine soja W05 v1" in the "Assembly" box.
Ignore genes with no hits
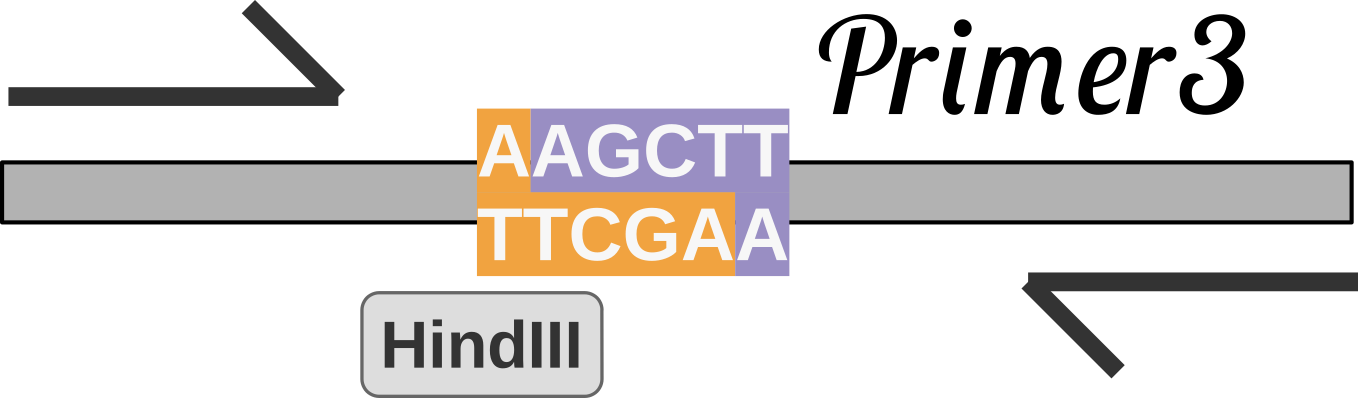
Basic Settings
5'-> 3' on plus strand

